Katrien Devos is Distinguished Research Professor with joint appointments in the University of Georgia’s departments of Crop & Soil Sciences and Plant Biology in the College of Agricultural & Environmental Sciences (CAES) and Franklin College of Arts and Sciences, respectively. Specializing in plant genetics, she also is a member of CAES’ Institute of Plant Breeding, Genetics & Genomics.
In this interview, she discusses her research into Panicum virgatum, a species native to North America commonly known as switchgrass, and the big role it could play in a more sustainable future.
Describe your work as it relates to biofuel production.
Our goal is to develop the perfect feedstock ideotype for bioenergy production. The feedstock, which is the starting material, could be a range of different materials. I work on switchgrass, which was selected [as a bioenergy crop] in the 1990s by the Department of Energy because it is high yielding, native to the U.S. and has high water-use efficiency and drought tolerance.
To develop the perfect ideotype, we need to understand the underlying genetics of traits that pertain to bioenergy production. Biomass yield is the foremost trait, but we’re also looking at composition, whether you have more sugars, more lignin, S/G lignin ratio, cell wall composition and so on—anything that has a direct or indirect effect on biomass yield and composition and, hence, fuel production.
UGA is heavily involved in the DOE-funded Center for Bioenergy Innovation (CBI) at Oak Ridge National Laboratory. What is your role?
I’m the lead for the Sustainable Switchgrass Domestication Team. One of my roles is to make sure there is close collaboration, coordination and synergy between the 12 team principal investigators, based at six institutions. For example, we have field trials in Georgia at the Iron Horse Farm in Watkinsville and the UGA Gibbs farm in Tifton, as well as at the University of Tennessee in Knoxville with the same switchgrass populations. We want to make sure we measure the same traits in very similar ways so we can combine the datasets to look at environmental effects.
We also coordinate research with other teams within CBI for specialized analyses. For example, we work with the Computational Biology Group to conduct genome-wide association studies and for explainable machine learning. We work with the Deconstruction Team to analyze the amount and composition of lignin, wax and sugars. We work with the Rapid Genetics Team, which does transformation in switchgrass, to validate genes we identify as involved in controlling traits of interest. We also work with the team that does economic modeling to determine the relative contribution of biomass yield and composition to fuel price.
What other research questions do you address, in terms of biofuels?
Our main interest is understanding the genetic control of traits important for bioenergy production. In addition to biomass yield, which is the primary determinant of biofuel cost, we’re also looking at a range of other traits. One is leaf wax, which gives the leaves of lowland switchgrass a bluish tint. Switchgrass occurs in two main ecotypes, upland and lowland. Genetically, they’re quite different, but you can cross them, which allows us to genetically map ecotypic differences such as wax.
Once we identified the gene that controls wax production, we worked with [Distinguished Research Professor] Wayne Parrott’s group, who is part of the Rapid Genetics Team in CBI, to knock that gene out in the waxy genotype, which resulted in plants with glossy, green leaves. Now we have these genetically identical plants that differ by one gene, the wax gene. How will this affect fermentation or deconstruction for bioenergy production?
Another consideration is sustainability in the field—if you remove the leaf wax, will that increase or decrease disease susceptibility? There is some evidence that fungal spores adhere better to leaves that contain wax. Studies on the effects of leaf wax on disease resistance are done in collaboration with Bochra Bahri, a plant pathologist and CBI member at the UGA Griffin Campus. In summary, we are looking at different aspects of how genetic changes affect bioenergy production, and how to harness these changes to improve yield as well as deconstruction of the plant.
How much trial and error is involved in this kind of genetic manipulation and selective breeding?
It depends on the trait. Sometimes, the results can be quite unexpected. When knocking out the wax gene, we got a plant that had glossy leaves, which we expected, but it also had other characteristics [that we didn’t expect] such as more tillers and reduced plant height. At this point, we have no explanation for this. Often, we think too simplistically, that if we knock a gene out, it will only affect one trait. But we know very well it’s going to have a knock-on effect on other traits, and we can’t always predict that.
Another example is flowering time. Flowering time is a good predictor of biomass, because the later a switchgrass plant flowers, the longer the vegetative growth period, typically resulting in more biomass. A graduate student in my group, Soyeon Choi, identified gene variants that control or contribute to differences in flowering time between upland and lowland accessions. In model systems, the functional gene variant induced earlier flowering. We are currently testing whether introducing this variant in lowland switchgrass will also accelerate flowering.
You were part of the team that mapped the switchgrass genome. How much of this work would be possible without that reference?
It would be difficult. You can get a long way with comparative genetics and genomics, using genomes of related species to study the genes and pathways that control traits in your crop of interest. But it isn’t perfect; the gene you’re after might be missing, or there might be genomic rearrangements that make it difficult to find the gene of interest. For example, we would never have found the wax gene using a comparative approach, because this is essentially a novel gene that was duplicated from elsewhere in the genome and then underwent fast evolution. So, both the gene and its location are unique to switchgrass.
Your ‘crop circles’ concept is an example of comparative genomics. Can you talk about that?
The crop circles show the relationship between genomes of different species. When I started my career, I worked on wheat, and we didn’t pay too much attention to what was going on in other crops. But then we started discovering that gene orders are highly conserved across species. Practically, this means that if you have a genome sequence from one species, you can use it to find causal genes for traits in another species.
For example, if you know a trait is located on chromosome 3 in wheat, and this region of chromosome 3 in wheat corresponds to chromosome 1 in rice, you can use that relationship to mine the rice genome sequence, which was the first monocot sequence to be generated, for the gene underlying your trait of interest in wheat.
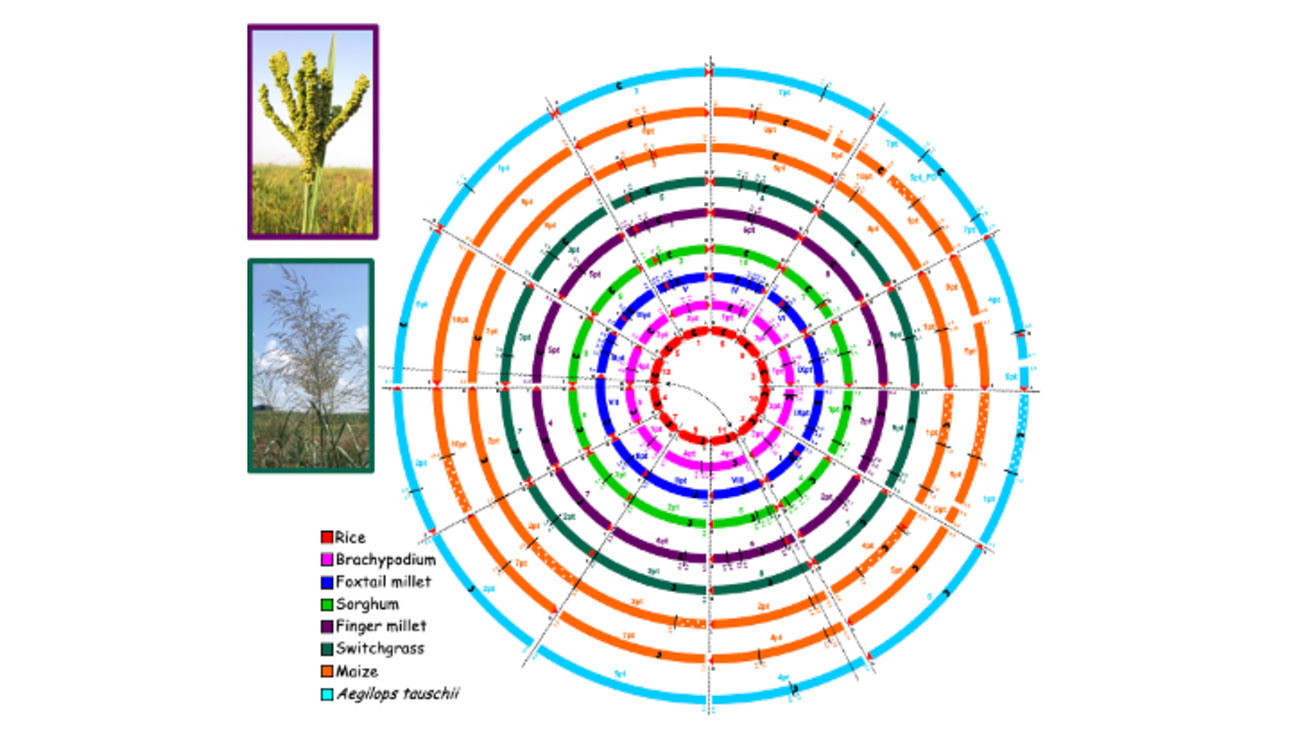
Earlier in her career, Devos and colleagues created a visual reference to help map relationships between genomes across different plant species. While not its original name, fellow researchers looked at the diagram and called it “Crop Circles.” The name stuck.
I was part of one of several groups that pioneered comparative studies in plants, and the crop circles compile all this information in an easy interpretable format. Let me give a practical example. Some groups worked on comparisons between rice and wheat, while others studied sorghum and maize in a comparative context, or maize and wheat. We pulled all this information together, and because side-by-side comparisons are not practical to display relationships between multiple species, we decided to use a circle format.
We originally called it comparative grass genome relationships, but the diagram quickly became known as the Crop Circles. And that name stuck.






